The Rye Laboratory
Department of Biochemistry and Biophysics
Protein disaggregation by molecular chaperones
The dark side of protein folding.
The misfolding and aggregation of essential cellular proteins is a fundamental problem for all living organisms. Aggregation of even non-essential proteins can lead to debilitating diseases like type II diabetes, Alzheimer’s, Huntington’s and Parkinson’s diseases.
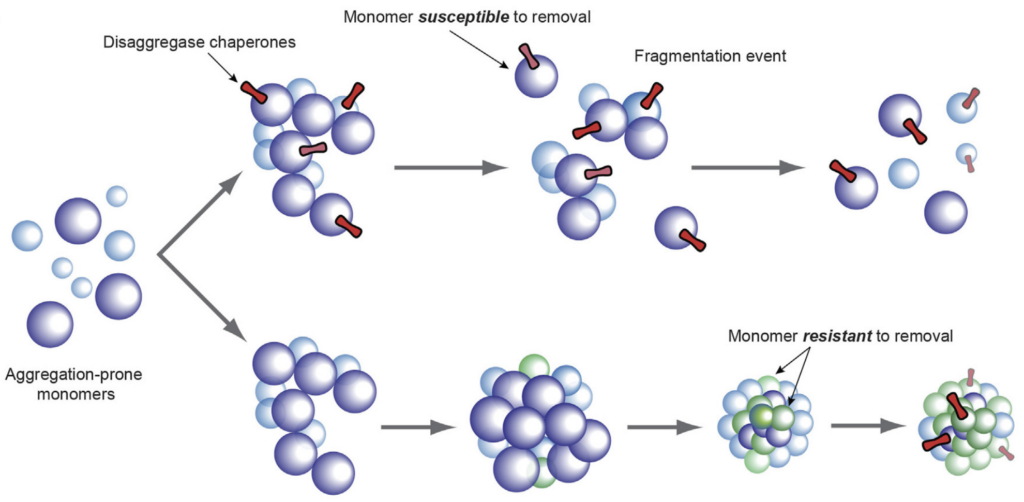
Importantly, protein folding and aggregation are heavily influenced by the cellular protein quality control machinery, involving networks of molecular chaperones. Precisely how different chaperone systems cooperate to dismantle and reactivate aggregated proteins, and how molecular chaperone action affects disease progression, is not well understood.
The difficulty of stydying protein disaggregation.
The investigation of protein disaggregation by molecular chaperones presents a serious technical challenge. In general, an aggregating protein can rapidly populate a heterogeneous mix of assembled states that span several orders of magnitude in size.
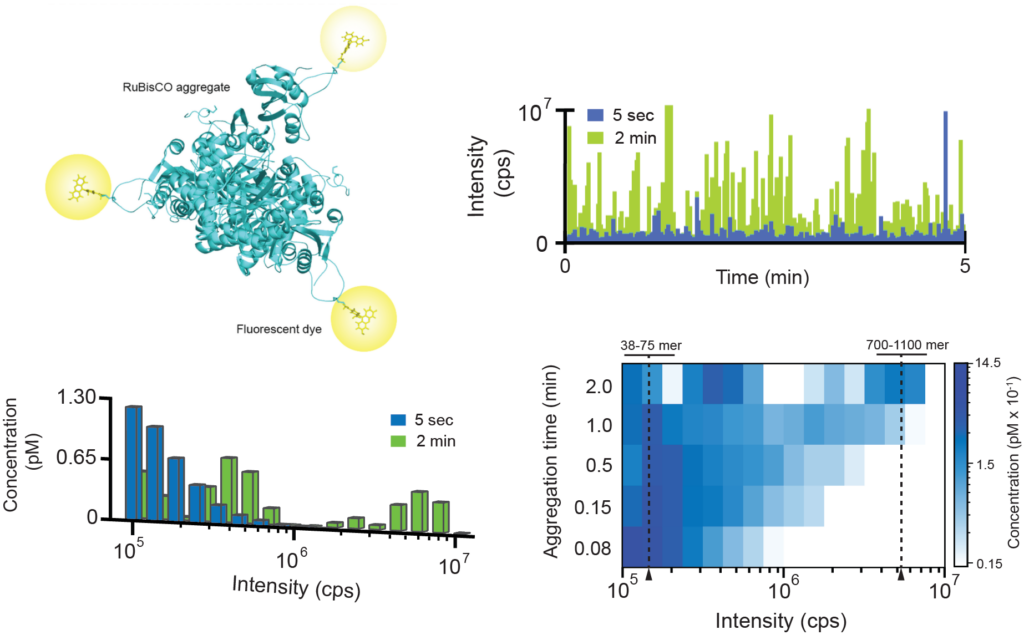
We are using a variety of approaches to study the disaggregase systems of both bacteria and yeast, including various fluorescence techniques like single molecule FRET, mass spectrometry and cryo-electron microscopy. In one approach, we employ fluorescently-labeled, aggregation-prone proteins like RuBisCO with single particle BAS measurements to measure the population-resolved kinetics of protein aggregate formation and disassembly by molecular chaperones.
How do molecular chaperone networks disassemble protein aggregates?
Interestingly, different organisms organize their protein disaggregation machinery in different ways, with most bacteria, many unicellular eukaryotes and plants employing a core bi-chaperone network composed of an Hsp70 system coupled to an Hsp100 system. By contrast, higher eucaryotes appear to exclusively employ an Hsp70-based disaggregase system.
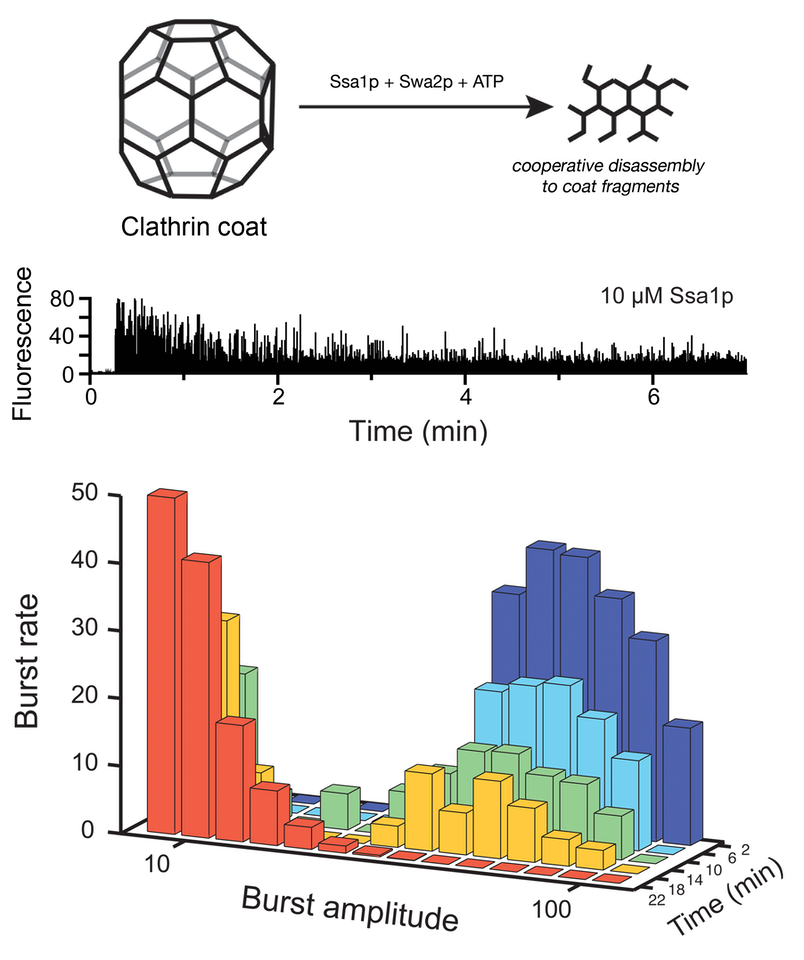
Hsp70 disassembly of clathrin coats
Hsp70/Hsp100 disaggregation
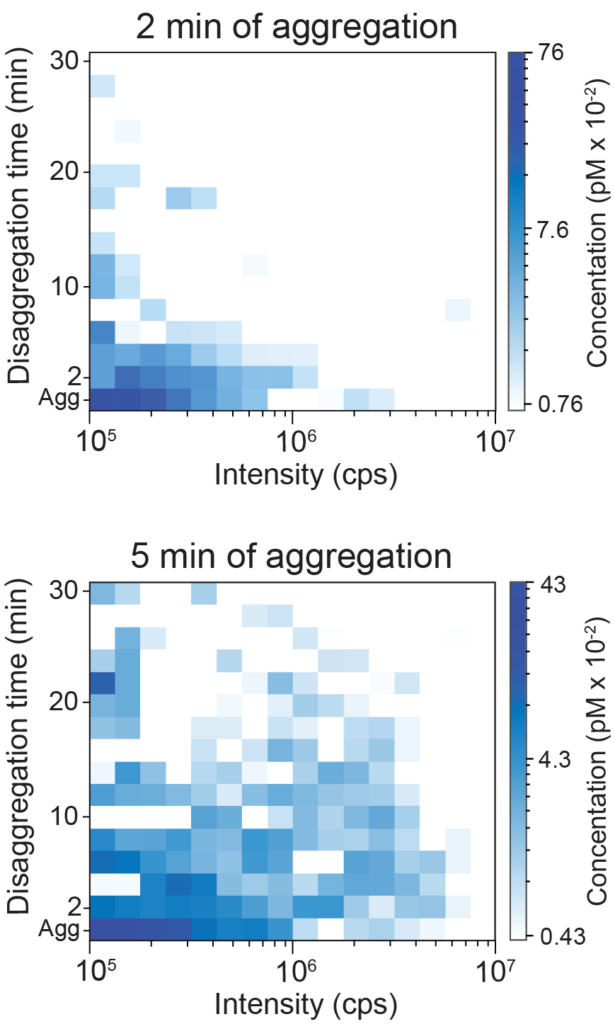
We have used BAS to examined the disassembly of fluorescent clathrin coats by a yeast Hsc70 molecular chaperone system consisting of Ssa1p and Swa2p (left). Analysis of the decay behavior of the large fluorescent bursts reveals that coat disassembly is highly cooperative. See JBC, 288: 26721 (2013). We have also used BAS to examined the disassembly of RuBisCO aggregates by the bacterial Hsp70/Hsp100 bi-chaperone disaggregase (right). Remarkably, aggregate particle size has only a minor impact on the rate at which aggregates can be taken apart. See Frontiers Mol Biosci, 9: 915307 (2022).